Ruth Young, Rhian White, Ilaria Zanetti, Naomi Cox, Erik Waskiewicz, Andrew Senior, Jenny Waizeneker and Rogan Vale
Cardiff and Vale University Health Board
Project Background:
In 2021 cancer caused ~25% of all UK childhood deaths (Children’s Cancers | Cancer Research UK). Current genetic testing for childhood cancers in Wales can include multiple genetic tests and invasive procedures.
Whole Genome Sequencing (WGS) – DNA sequencing method for whole genome analysis. One test can detect a variety of genomic abnormalities including tumour and familial (cancer predisposition genes) abnormalities through paired testing of tumour and germline DNA (Figure 1). (Nakawaga et al., 2015).
There is evidence that WGS provides more accurate diagnosis, prognosis and access to more treatment options for childhood cancer than current standard of care genetic tests (Trotman et al., 2022).
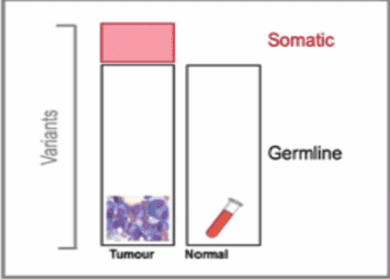
Graphical demonstration of WGS capability to detect tumour (somatic) and germline variants through paired testing of tumour and germline DNA.
Project Aims:
- Validate a whole genome sequencing (WGS) pipeline for paediatric oncology patients at diagnosis, relapse, progression or where all standard of care treatment options are exhausted (<25years)
- Develop a clinical and laboratory pathway that utilises a PCR free WGS testing method for paediatric oncology samples and matched germline samples.
Project Outcomes:
Procedures in place for sample referral, patient consent, laboratory receipt, sample storage, sample culturing, DNA extraction and sequencing from required samples for WGS for paediatric oncology patients.
- 44/45 sets of patient samples passed DNA extraction quality control parameters for WGS.
- 25/27 sequencing runs met quality parameters (One run repeated twice).
- 10/12 sequencing runs met initial bioinformatics quality parameters (One run repeated twice).
All SNVs (single nucleotide variants) with greater than 5% VAF (Variant allele frequency) detected in the cancer cell line using TSO500 DNA in house cancer NGS panel were also detected using WGS.
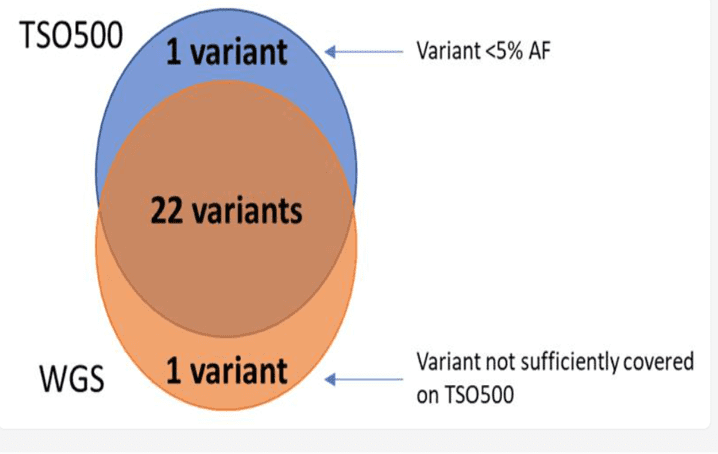
Cancer cell line sample: Venn diagram comparison of variants detected using TSO500 in house cancer DNA NGS panel and WGS.
Project Impact:
An in house WGS service for paediatric oncology patients is possible. DNA extracted from paediatric oncology samples is of sufficient quality for initiation of WGS.
First line WGS testing for paediatric oncology patients could replace the majority of separate standard of care genetic tests and provide greater chance of more accurate diagnosis, treatment and prognosis in the first instance. This could prevent repetitive genetic testing and limit sampling procedures experienced by children.
A paediatric oncology WGS service would be utilised by approximately 80 Welsh patients per year.
Genomics Partnership Wales – Patient and Public sounding board feedback – The sounding board were generally positive about using WGS technology for cancer patients and the patient benefit it could bring.
‘Information is power’





